Cell fate emerges from complex interplay between processes occurring on many biological scales. At CMCF, we study the impact of this complex regulation to address several large open questions in biology. Our current projects focus on different (but interlinked) components of cell fate, which all seek to identify the emergent complexity of multiscale systems using experimental and mathematical methodologies.
The form and function of tissues and organs emerge out of cell-cell interactions. Cell interaction dynamics take place on a variety of temporal and spatial scales, and reflect processes — diffusion, migration, force production/sensing, growth, and proliferation — considerably more complex than the basic mass-action chemistry that underlies gene regulatory networks within cells. Understanding how tissues and organs work means developing models that integrate such processes with spatiotemporally resolved data on gene expression and cell lineages. Doing this requires obtaining data from individual cells, a great many at a time, a problem that has long hindered the development of predictive and explanatory tissue- and organ-scale models. Recent technological breakthroughs have, however, changed this. For example, single cell RNA sequencing (scRNA-seq) now makes it possible to comprehensively probe gene expression in thousands of individual cells simultaneously. We investigate important dynamical processes such as the control of size and form, which can be achieved through the regulation of cell proliferation. Such control is common to many tissues and organs, yet poorly understood. It may manifest as homeostasis, regeneration, or rapid and accurate development. It is known that feedback (mechanical or chemical) is essential: we seek a detailed mechanistic understanding of the details of such feedback.
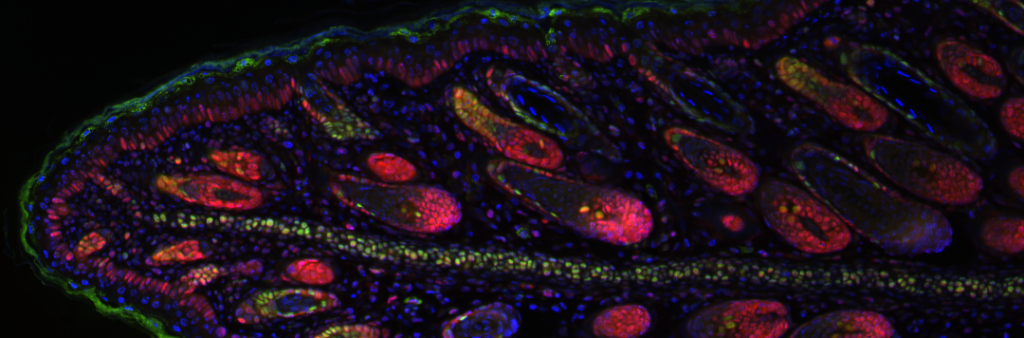
During development, cell fates are often specified in noisy and dynamic three-dimensional environments, which cells must navigate through, e.g. by migration. Examples include the formation of segments of the hindbrain and the pharyngeal arches, precursors of the jaw and larynx; these are largely composed of cranial neural crest cells, arising from distinct segmental positions in the hindbrain and migrating in streams through the head. While prevailing theories suggest that premigratory neural crest cells are pluripotent, relying on their migratory environment for fate specification, some lineage tracing studies have hinted at earlier pre-specification. It remains elusive when, where, and how neural crest cells acquire fate identities and robustly migrate to correct locations despite gene expression fluctuations within each cell, and fluctuations due to environmental signals. We study these stochastic dynamics in developmental systems. Heterogeneity is critical to robust environmental responses during migration. We speculate that temporal heterogeneity may present greater opportunities for fate specification, in a mechanism whereby persistent signals may be differentially interpreted by cells to regulate their migration paths and eventual fates.
Cell fate can be described in terms of transcription factor (TF) networks, in which dynamic attractors correspond to cell types. Yet in the lab and the clinic, our ability to drive cells towards desired differentiation paths remains poor. This is, in large part, because we lack a quantitative understanding of these networks. A major hurdle to such an understanding is the “chicken-and-egg” nature of TF–chromatin landscape interplay: TFs help coordinate chromatin remodeling across the genome; accessibility of genomic regions to TFs is in turn governed by chromatin. Given this complexity, what principles are exploited to control cell transitions from one phenotype to another? One possibility is that cells primarily modulate TF levels, i.e. action in trans; the alternative, action in cis, is that the the DNA is itself regulated, via changes in methylation, histone modifications, or nucleosome positioning. We are studying cis-regulation as a major modulator of phenotype transitions, mediated by dynamic DNA methylation. The necessity of regulation in cis could resolve several puzzles, such as why “global” epigenetic modifiers, such as DNA methyltransferase inhibitors, have lineage-specific effects, and moreover, why TF-TF interactions are cell-type-, chromatin-, or otherwise context-dependent.
