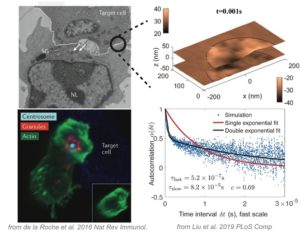
Computational Fluid Dynamics of Immune Cells (Jun Allard)
T Cells are immune cells responsible for surveilling the body for diseased cells – but sometimes they go awry, for example producing “cytokine storms” that have killed tens of thousands of humans in the past few months. To operate effectively, T Cells must push up against diseased cells and secrete toxic chemicals that are only received by the diseased cell, and not bystander cells. It has been hypothesized that the chemicals are released through a fluid-filled bubble called a secretory cleft. We previously used computational fluid dynamics to study the initial stages of T Cell contact [Liu, Read, Lowengrub, Allard 2019 PLoS Comp] applicable to CD4+ T Cells, which do not form secretory clefts. Here, we will use computational fluid dynamics to study the secretory cleft in CD8+ T Cells. How does the T Cell maintain a tight contact with the diseased cell, so no or few toxins leak out? The project involves the following. Tools: high-performance computing, Docker containers, bash and C++. Scientific topics: biophysics, fluid dynamics, immunology, cell biology.
COVID19: Data And Analysis (Natalia Komarova)
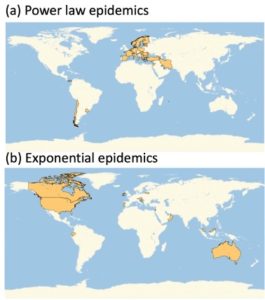
In this project we will be using data that are available on the growth of COVID19 pandemic in different countries, different US states, or different California counties. We will fit mathematical models to data, and try to figure out how infection spreads and what measures are effective in slowing it down. The attached figure is from our preliminary analysis of COVID19 initial growth data, which allowed us to classify different countries by the type of growth law that was governing the expanding epidemic. In the new projects we will use data if social distancing, networks and interactions to understand the spread of the disease and suggest best measures for its control.
Stochastic Models of Kidney Stone Formation (German Enciso)
Metabolic Symbiosis in Tumors (John Lowengrub)
A hallmark feature of many cancers, and an enduring puzzle of biology since its discovery in the 1950’s, is a phenomenon called the Warburg Effect. Here, cancerous cells gain access to oxygen, but then shift their metabolism to aerobic glycolysis, away from (oxygen-using) oxidative phosphorylation (OXPHOS) to favor glycolysis, despite the availability of sufficient levels of oxygen. This is especially intriguing since glycolysis is much less efficient than OXPHOS in producing energy. However, it is increasingly recognized that glycolysis is not a singular choice for all cells in a tumor; OXPHOS modes of metabolism may be dominant to some cells but not others. The proposed outcome of this heterogeneity is that cooperation between these two groups of cancer cells can maximize the delivery and consumption of nutrients and minimize the environmental stresses that are imposed on a tumor. This is known as metabolic symbiosis. For example, glycolytic cells produce lactate that can be used as fuel by cells using OXPHOS. Figure 1 shows metabolism under varying oxygen concentrations.
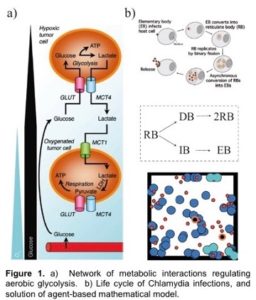
In this project, students will develop deterministic and stochastic population-based and individual-based mathematical models to determine conditions under which metabolic symbiosis is not just a manifestation of tumor heterogeneity, but is a fundamental aspect of tumor survival. That is, students will determine conditions and parameter ranges for which tumors that display metabolic symbiosis have an enhanced fitness compared to those in which there is metabolic homogeneity. The population-based modeling will use ordinary and stochastic differential equation models while the individual-based modeling will use CompuCell 3D, which is based on the Cellular Potts approach, as the primary modeling tool. We have extensive experience using these approaches. Model predictions will be compared to results in the experimental literature.
The mathematical challenge lies in developing a model that, on the one hand, contains all the relevant biological features needed to compare with experimental data, and, on the other, is simple enough to provide insight into the underlying mechanism of spatial heterogeneity.
Deep Learning Human Hair Follicle Dynamics (Qing Nie and Max Plikus)
Hair loss is a problem that affects millions of people around the world. One promising treatment is to induce hair growth in waves, but this requires differentiating between different stages of the hair growth cycle on humans. We use a non-invasive skin imaging modality called OCT (pictured below) to find imaging parameters in order to make these hair cycle staging decisions. The goal of this project is to help process these 3D images and use machine learning to differentiate between the stages of hair growth. This would involve some classical image analysis as well as exposure to deep learning for image processing.
TAs: Sean Kuan (Plikus lab) and Eric Chang (Nie lab)
Site Specific Differences in Development of Dermal Pericytes (Bogi Andersen and Qing Nie)
Volar skin, the thickened skin found on the palms and soles, is marked by its increased mechanical strength, and its ability to regularly bear high loads. Compared to non-volar skin, it is marked by the increased thickness of the epidermis and interdigitation between the dermis and epidermis, which are thought to lend to its increased toughness. Although these site specific differences have been well studied previously, our human single cell RNA-seq (scRNA-seq) data, comparing volar (palm and sole) to non-volar (hip) skin, provides the opportunity to more comprehensively study these differences in a variety of cell types. Pericytes, which are mural cells surrounding blood vessels, have recently been shown to play a role in skin regeneration. Using our scRNA-seq data, we can identify whether heterogeneity exists within the these pericytes, and determine whether certain pericyte subclusters are enriched in volar skin. Additionally, we can attempt to identify pericyte progenitors, mesenchymal stem cells, in our dataset and construct a differentiation trajectory that will trace from these earliest progenitors to the differentiated pericytes. Finally, we can work to identify regulators of pericyte gene expression, and determine which regulators are conserved and which differ between volar and non-volar skin.
TAs: Julie Wiedemann (Andersen lab) Yutong Sha (Nie lab)
Noise in Genomic Sequencing (Elizabeth Read)
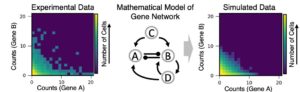
In recent years, new technology in genomic sequencing has enabled researchers to measure the expression of many thousands of genes simultaneously within individual cells. In a population of cells (extracted, for example, from animal or human tissue), these single-cell-level gene expression measurements appear very noisy—that is, they show big differences between individual cells. This project asks the question: How much of the noise in these data is useful noise? In other words, do cell distributions contain hidden information about underlying regulatory relationships between genes, and therefore about the gene networks that help determine an individual cell’s identity? Inferring such gene networks is an ongoing challenge for fundamental biological discovery and applied biotechnology. To address these questions, we will use mathematical modeling and simulation of small gene regulatory networks with defined characteristics. Noise properties of simulated networks will be compared to those of experimental data in order to determine the limits of what the noise properties can and can’t tell us about the underlying networks.
Mapping Electrostatic Impacts of Antiviral Drugs on Viral RNA Polymerase in SARS-CoV-2 (Jin Yu)
RNA-dependent RNA polymerase (RdRp) is a critical enzyme responsible for replication and transcription in RNA viruses such as the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) that causes current coronavirus disease 2019 (COVID-19) pandemic.
Investigating the mechanism of action of RdRp from SARS-CoV-2 (Cov-RdRp) offers promise for development of new antiviral therapeutics and for understanding the emergence of drug resistance. Our computational biophysics lab (https://sites.uci.edu/jinyulab/) employs molecular modeling and simulation techniques to investigate the structural dynamics underlying the Cov- RdRp function (Fig 1 left). For example, Remdesivir, a prodrug of a nucleotide analog, will be utilized to interrogate RdRp fidelity and better understand the basis for its antiviral activity (Fig 1). Our computation is currently supported by the COVID-19 HPC Consortium, using the world leading supercomputer Summit from the Oak Ridge National Lab.
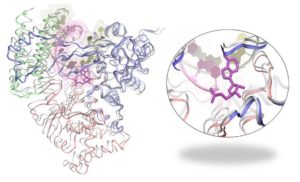
Fig 1: The viral RdRp protein enzyme from SARS-CoV-2 (left) and a close view of its active site docked with a nucleotide analog from the anti-viral drug compound Remdesivir
In this project, we focus on solving the Poisson-Boltzmann equation of the antiviral drug bound Cov-RdRp system, in order to map electrostatic impacts of various drug compounds on the Cov-RdRp protein. We will use software with the specialized Poisson-Boltzmann solver in combination with molecular graphics and visualization tool, molecular docking and force field optimization, and molecular dynamics simulation packages to accomplish the computational tasks. The key scientific question remains open and challenging for this project is, whether and how various potential drug molecules impact on the Cov-RdRp functioning electrostatically upon binding/incorporation.
See Dr. Yu’s project intro video here.

